Aim of this project
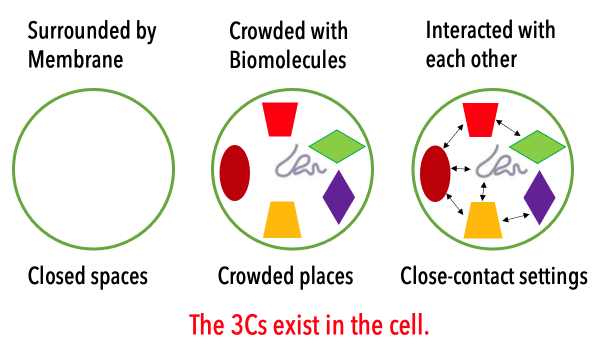
The cell is covered with the cellular membrane (Closed spaces). The inside of the cell is highly crowded with many biomolecules (Crowded places), which weakly interact with each other (Close-contact settings). In this project, we aim to understand how the cellular environments affect the structure and function of each protein and nucleic acid in the cell. Another essential question to be solved is how the cellular environments are composed of proteins and other biomolecules in the 3Cs condition. In contrast to the conventional biology, we rather focus on the roles and reconstructions of the cellular environments. For this purpose, we need to integrate experimental biology with computational biophysics for elucidating dynamic structures and weak molecular interactions in the cells.
Research groups and collaboration
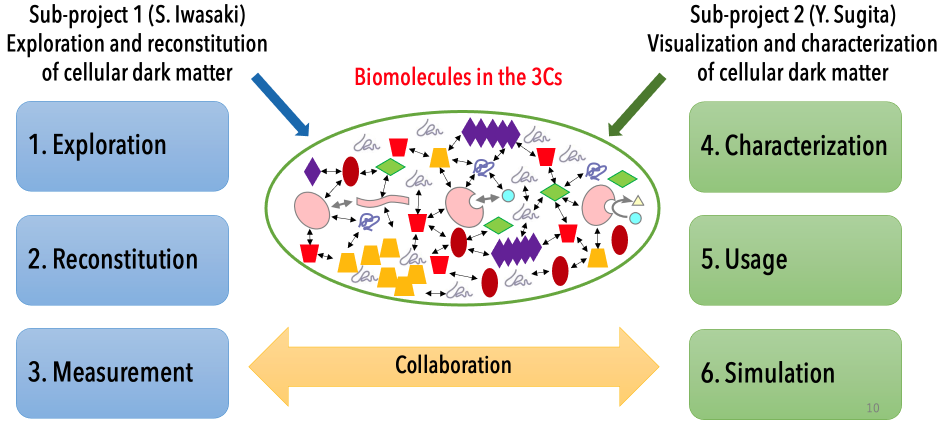
Sub-project 1: Exploration and reconstitution of intracellular environments
Hero (Heat-resistant obscure protein) is one of the structure-function unknown proteins in the cellular environments. This class of proteins shows heat resistance in its function and is disordered in the structure. Hero can avoid the disorder or disfunction of other proteins in the crowded cellular environments and therefore, Hero itself has essential functions as a part of the cellular environments. We utilize Ribosome profiling method and mass spectroscopy to explore Hero proteins and their target molecules in mammals and plants. We also study other essential proteins that form the cellular environments and regulate structures and functions of other biomolecules in the cell. We also utilize cryo-electron tomography to visualize the cellular crowded environments directly. In this way, we investigate various biomolecules whose structures and functions are currently unknown while important to affect cellular functions. This project will be conducted with multidisciplinary strategies using various techniques including bioinformatics, genetics, biochemistry, microscopy, tomography, and biophysics.
Sub-project 2: Visualization and characterization of intracellular environments
We analyze dynamics structures of various biomolecules whose structures and functions are currently unknown in the cellular environments mainly via structural biology and computational biophysics. We aim to understand the effect of crowded cellular environments on individual proteins and nucleic acids as well as the molecular mechanisms for how proteins and other biomolecules construct the crowded cellular environments. Intrinsically disordered proteins, flexible RNAs, and their weak and nonspecific interactions are key features to be examined in detail in the sub-project. We utilize solution and in-sell NMR spectroscopy, cryo-electron microscopy and tomography, and single molecule measurements as experimental techniques. The experimental studies are tightly collaborated with the latest computational methods, namely, multi-scale molecular dynamics simulations, machine learning, and data-assimilation. By integrating the experimental and computational techniques, we propose new strategies in life science for tackling difficult problems.
